Plan of attack
Source of data
Let’s do a quick PubMed search. Copy the following search terms and search on PubMed. Search terms: (covid 19) AND ((“2020/08/02”[Date - Publication] : “3000”[Date - Publication]))
Alternatively you can find the search link here.
With the above search terms, we can retrieve 601 COVID-19 related records that were indexed in PubMed on August 03, 2020.
We can now open these records using our citation manager: EndNote, Mendeley, etc.
Since the file is in RIS format, we must convert it to BibTeX so that we can benefit from Bibliomterix package.
I will come back in the future in another blog post to show you on how to convert RIS file to BibTeX or converting from CSV/XSL reference file to EndNote-readable format. For today, let’s just use the converted COVID19 BibTeX file. We can load convert our BibTeX file using either bibliometrix and then bib2df package.
Let’s first use bibliometrix and then bib2df package to convert our COVID19 BibTeX file to data frame.
Initialize
Set working directory
Load necessary packages
Import the data to R: Covert the BibTeX file to data frame
covid19_bibanalysis <- convert2df("covid19.bib", dbsource = "isi", format = "bibtex")
Converting your isi collection into a bibliographic dataframe
Warning:
In your file, some mandatory metadata are missing. Bibliometrix functions may not work properly!
Please, take a look at the vignettes:
- 'Data Importing and Converting' (https://www.bibliometrix.org/vignettes/Data-Importing-and-Converting.html)
- 'A brief introduction to bibliometrix' (https://www.bibliometrix.org/vignettes/Introduction_to_bibliometrix.html)
Missing fields: ID C1 CR
Done!#As usual, isnpect the data
dim(covid19_bibanalysis) #601 records, 23 variables[1] 601 25glimpse(covid19_bibanalysis) #Inspect the structure of data, variable names, etcRows: 601
Columns: 25
$ AU <chr> "GELZINIS TA", "COCCIA M", "ATAGUBA OA;ATAGUBA J~
$ DE <chr> NA, "AIR POLLUTANTS;AIR POLLUTION;BETACORONAVIRU~
$ AB <chr> NA, "THIS STUDY HAS TWO GOALS. THE FIRST IS TO E~
$ BO <chr> "JOURNAL OF CARDIOTHORACIC AND VASCULAR ANESTHES~
$ DI <chr> "10.1053/j.jvca.2020.05.008", "10.1016/j.scitote~
$ institution <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, ~
$ SN <chr> "1532-8422 (ELECTRONIC)", "1879-1026 (ELECTRONIC~
$ SO <chr> "JOURNAL OF CARDIOTHORACIC AND VASCULAR ANESTHES~
$ LA <chr> "ENG", "ENG", "ENG", "ENG", "ENG", "ENG", "ENG",~
$ month <chr> "SEP", "AUG", "DEC", "SEP", "DEC", "NOV", "SEP",~
$ PN <chr> "9", NA, "1", NA, "1", NA, NA, "1", NA, NA, "3",~
$ PP <chr> "2328--2330", "138474", "1788263", "312--321", "~
$ pmid <chr> "32406428", "32498152", "32657669", "32546875", ~
$ TI <chr> "THORACIC ANESTHESIA IN THE CORONAVIRUS DISEASE ~
$ VL <chr> "34", "729", "13", "117", "9", "110", "83", "9",~
$ PY <dbl> 2020, 2020, 2020, 2020, 2020, 2020, 2020, 2020, ~
$ DB <chr> "ISI", "ISI", "ISI", "ISI", "ISI", "ISI", "ISI",~
$ JI <chr> "J. Cardiothorac. Vasc. Anesth.", "Sci. Total. E~
$ J9 <chr> "J. Cardiothorac. Vasc. Anesth.", "Sci. Total. E~
$ TC <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, ~
$ CR <chr> "none", "none", "none", "none", "none", "none", ~
$ C1 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, ~
$ AU_UN <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, ~
$ SR_FULL <chr> "GELZINIS TA, 2020, J. Cardiothorac. Vasc. Anest~
$ SR <chr> "GELZINIS TA, 2020, J. Cardiothorac. Vasc. Anest~Now bibliometrics analysis
results <- biblioAnalysis(covid19_bibanalysis, sep = ",") #create the object
options(width=100) #to determine width of the plot
s <- summary(object=results, k=10, pase=FALSE) #to present 10 most prominent authors, journals, keywords, etc
MAIN INFORMATION ABOUT DATA
Timespan 2020 : 2021
Sources (Journals, Books, etc) 206
Documents 601
Annual Growth Rate % -99.67
Document Average Age 3
Average citations per doc 0
Average citations per year per doc 0
References 1
DOCUMENT CONTENTS
Keywords Plus (ID) 0
Author's Keywords (DE) 269
AUTHORS
Authors 594
Author Appearances 601
Authors of single-authored docs 594
AUTHORS COLLABORATION
Single-authored docs 601
Documents per Author 1.01
Co-Authors per Doc 1
International co-authorships % 0
Annual Scientific Production
Year Articles
2020 599
2021 2
Annual Percentage Growth Rate -99.67
Most Productive Authors
Authors Articles
1 NA NA 3
2 BENTATA Y 2
3 DAUGHTON CG 2
4 HERMAN JA;URITS I;KAYE AD;URMAN RD;VISWANATH O 2
5 KAPOOR I;PRABHAKAR H;MAHAJAN C 2
6 TAHAN HM 2
7 ABD EL-AZIZ TM;STOCKAND JD 1
8 ABDELMAKSOUD A;GOLDUST M;VESTITA M 1
9 ABDULLAH MS;CHONG PL;ASLI R;MOMIN RN;MANI BI;METUSSIN D;CHONG VH 1
10 ABDULLAH S;MANSOR AA;NAPI NNLM;MANSOR WNW;AHMED AN;ISMAIL M;RAMLY ZTA 1
Authors Articles Fractionalized
1 NA NA 3
2 BENTATA Y 2
3 DAUGHTON CG 2
4 HERMAN JA;URITS I;KAYE AD;URMAN RD;VISWANATH O 2
5 KAPOOR I;PRABHAKAR H;MAHAJAN C 2
6 TAHAN HM 2
7 ABD EL-AZIZ TM;STOCKAND JD 1
8 ABDELMAKSOUD A;GOLDUST M;VESTITA M 1
9 ABDULLAH MS;CHONG PL;ASLI R;MOMIN RN;MANI BI;METUSSIN D;CHONG VH 1
10 ABDULLAH S;MANSOR AA;NAPI NNLM;MANSOR WNW;AHMED AN;ISMAIL M;RAMLY ZTA 1
Top manuscripts per citations
Paper
1 GELZINIS TA, 2020, J. Cardiothorac. Vasc. Anesth.
2 COCCIA M, 2020, Sci. Total. Environ.
3 ATAGUBA OA, 2020, Glob. Health Action
4 SIGALA M, 2020, J. Bus. Res.
5 LIU XH, 2020, Emerg. MICROBES \\& Infect.
6 LECHNER WV, 2020, Addict. Behav.
7 CAGLIANI R, 2020, Infect. Genet. Evol. : J. Mol. Epidemiol. Evol. Genet. Infect. Dis.
8 OKBA NMA, 2020, Emerg. MICROBES \\& Infect.
9 DONTHU N, 2020, J. Bus. Res.
10 VAN DORP L, 2020, Infect. Genet. Evol. : J. Mol. Epidemiol. Evol. Genet. Infect. Dis.
DOI TC TCperYear NTC
1 10.1053/j.jvca.2020.05.008 0 0 NaN
2 10.1016/j.scitotenv.2020.138474 0 0 NaN
3 10.1080/16549716.2020.1788263 0 0 NaN
4 10.1016/j.jbusres.2020.06.015 0 0 NaN
5 10.1080/22221751.2020.1766383 0 0 NaN
6 10.1016/j.addbeh.2020.106527 0 0 NaN
7 10.1016/j.meegid.2020.104353 0 0 NaN
8 10.1080/22221751.2020.1760735 0 0 NaN
9 10.1016/j.jbusres.2020.06.008 0 0 NaN
10 10.1016/j.meegid.2020.104351 0 0 NaN
Most Relevant Sources
Sources Articles
1 EMERGING MICROBES \\& INFECTIONS 61
2 THE SCIENCE OF THE TOTAL ENVIRONMENT 54
3 CHAOS SOLITONS AND FRACTALS 22
4 MEDICAL EDUCATION ONLINE 21
5 JOURNAL OF AFFECTIVE DISORDERS 18
6 JOURNAL OF CLINICAL ANESTHESIA 18
7 SEXUAL AND REPRODUCTIVE HEALTH MATTERS 14
8 NEUROLOGY(R) NEUROIMMUNOLOGY \\& NEUROINFLAMMATION 13
9 INFECTIOUS DISEASES (LONDON ENGLAND) 10
10 INTEGRATIVE MEDICINE RESEARCH 10plot(x=results, k=10, pause=FALSE) #plot the results
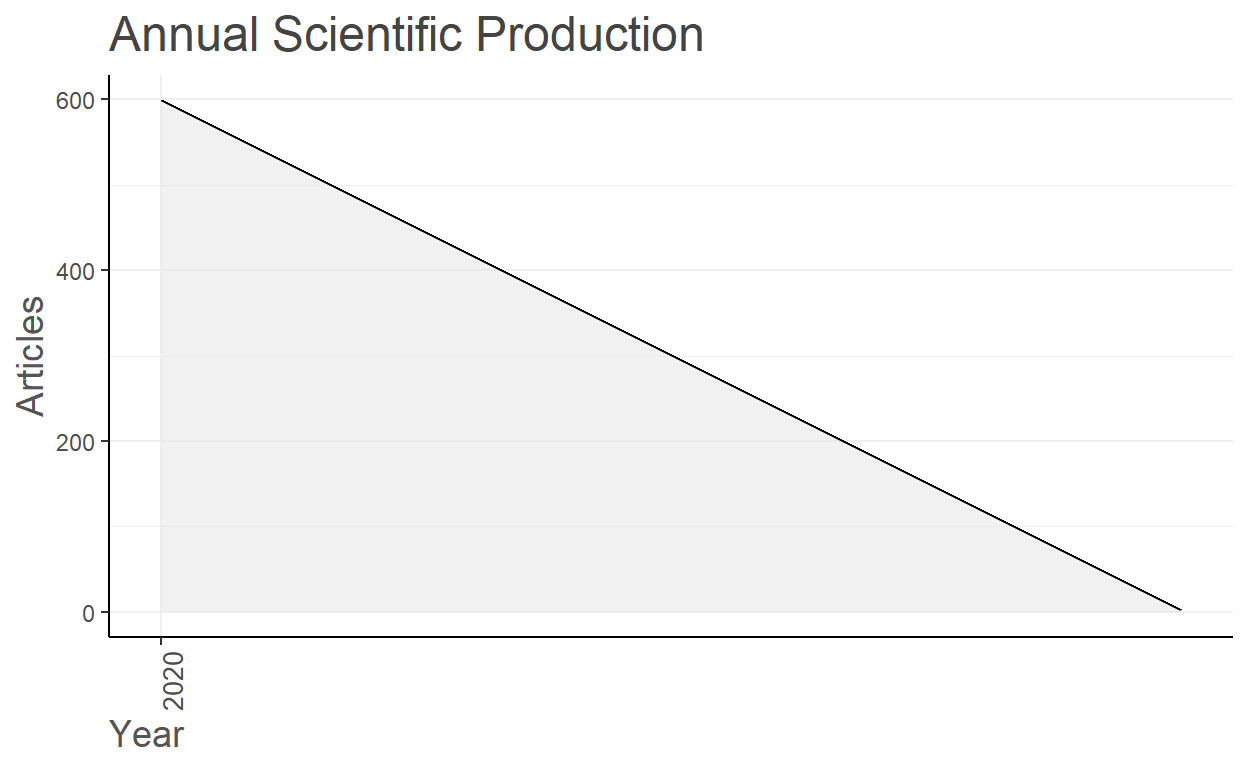
#since all of them are indexed in August 2020, this may not be useful analysis.
# Let's see sankey plots. But, before that we must remove missing values
threeFieldsPlot(covid19_bibanalysis, fields = c("AU", "DE", "SO"))Look at this plot. It maps Authors with keywords and with the journals. With 61 articles indexed in PubMed in one day, a journal called, EMERGING MICROBES \& INFECTIONS is leading the league.
## Now let's do some clustering
# Let's just use key words
cs <- conceptualStructure(covid19_bibanalysis, field="DE",
method="CA", minDegree = 4,
stemming=FALSE, labelsize = 10, documents=2) 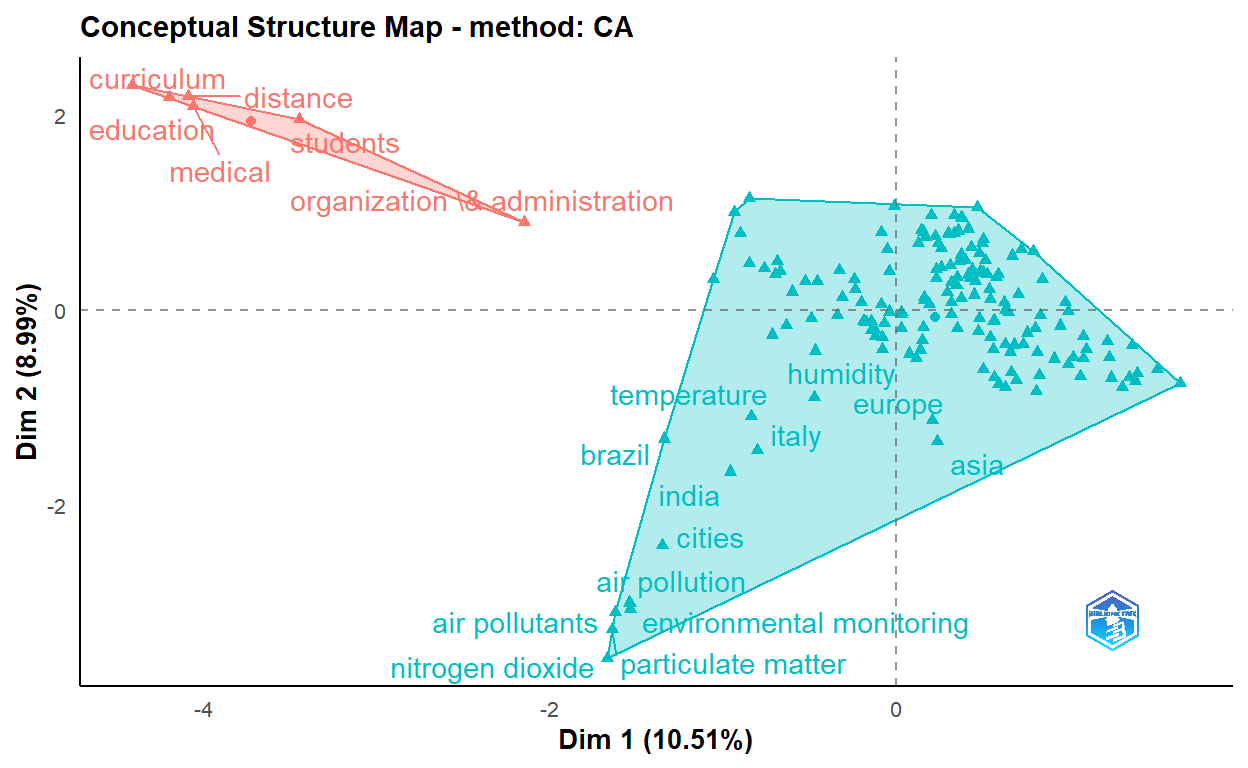
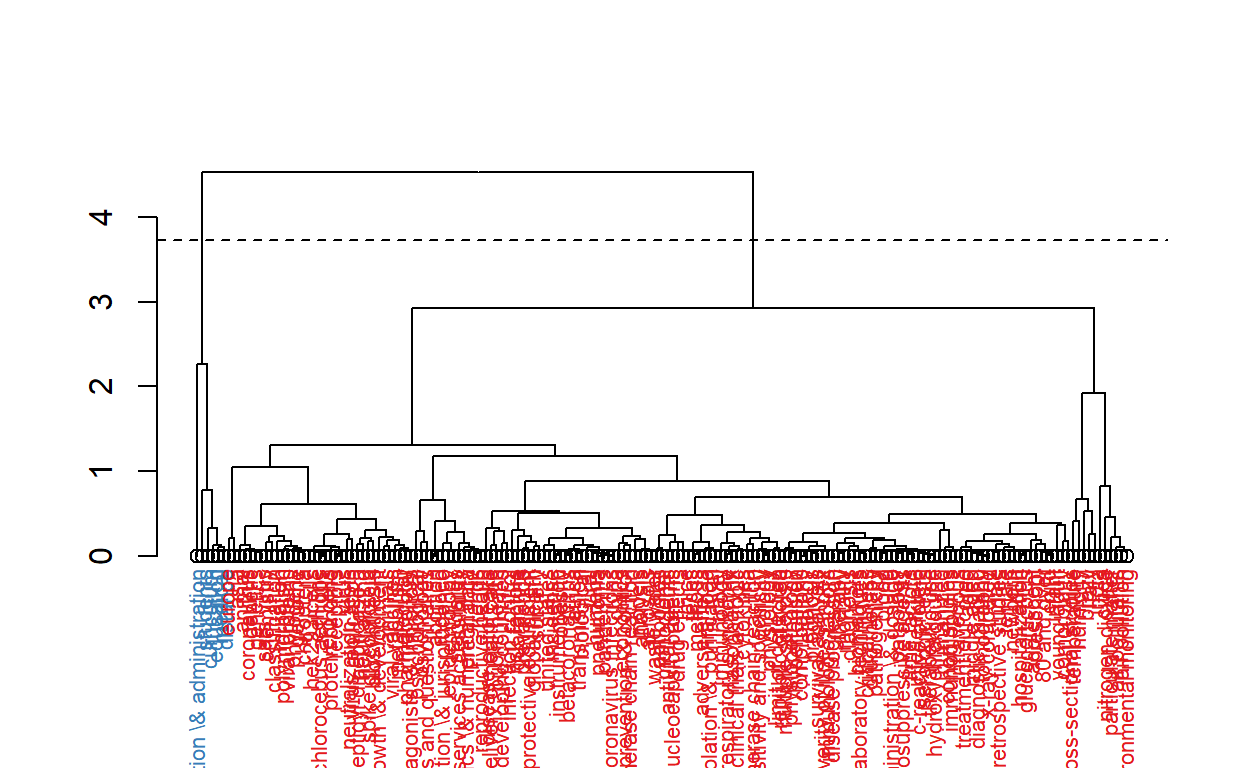
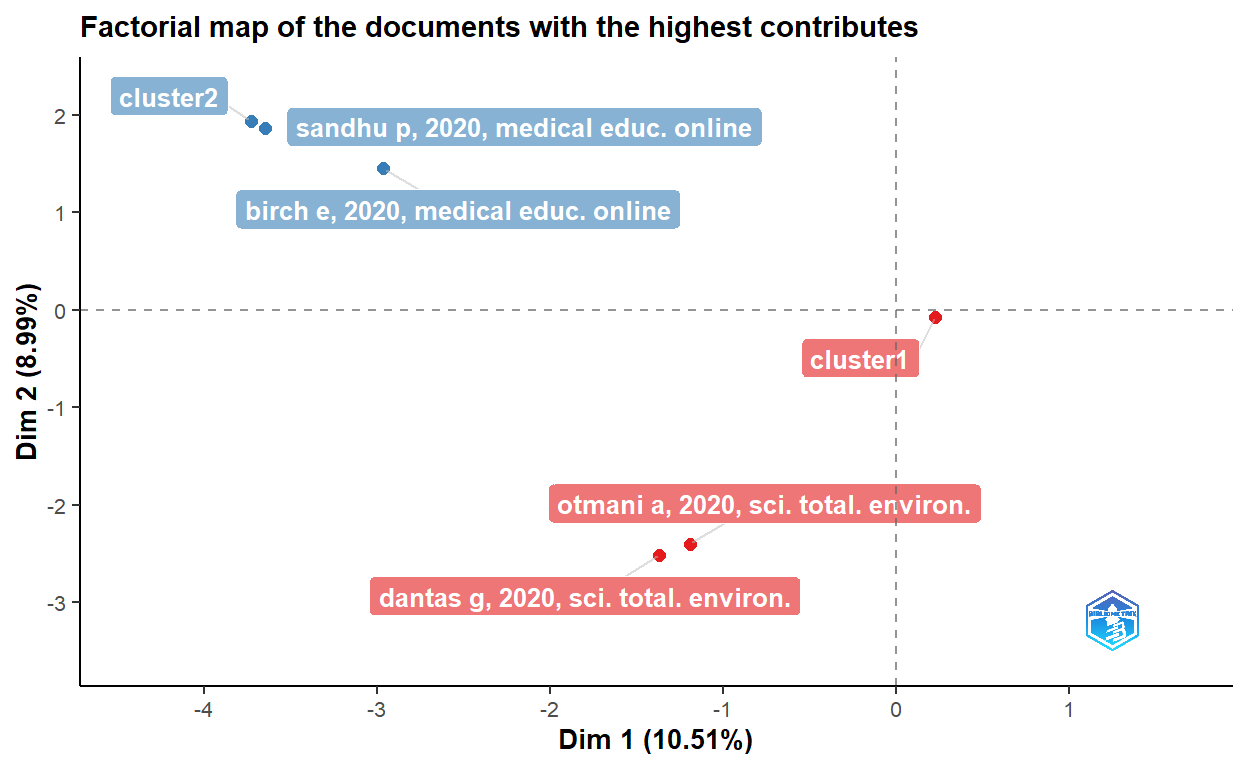
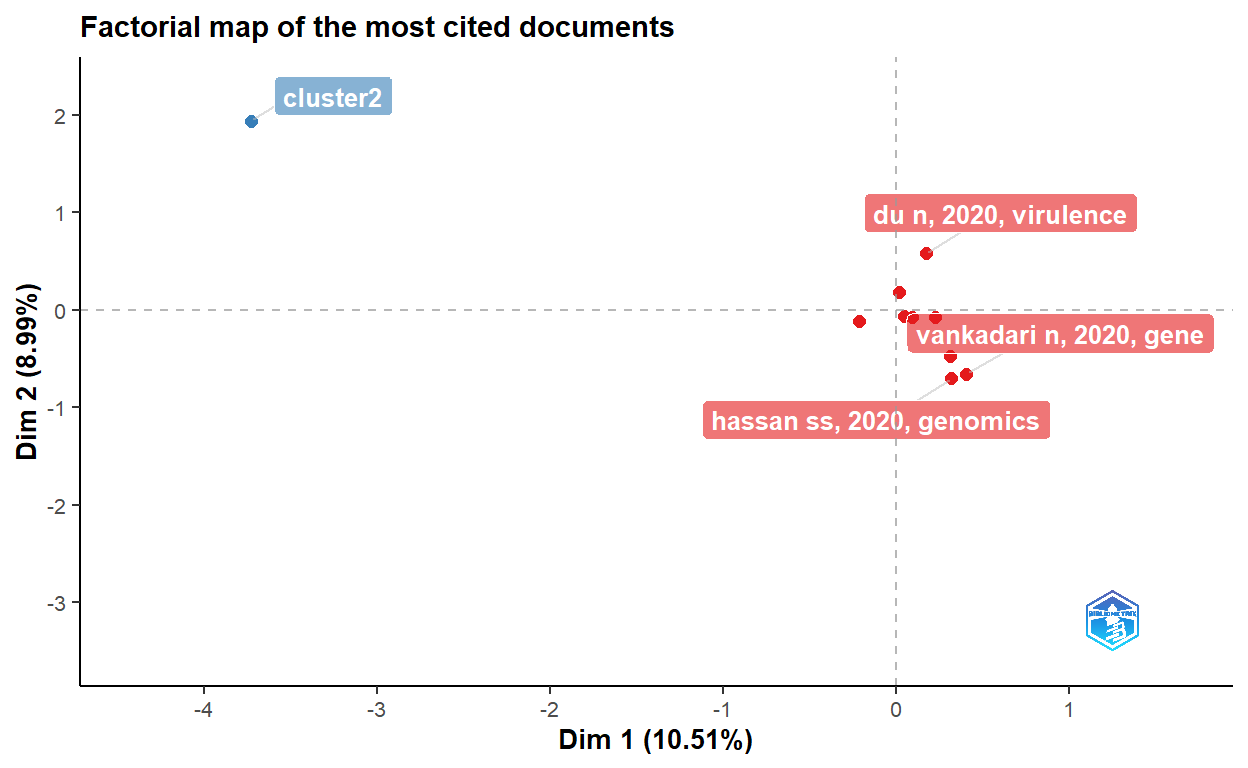
# Let's now use Akey words
# is abit dense to vizualize it here. It is very dense.
csAB <- conceptualStructure(covid19_bibanalysis, field="DE",
method="CA", minDegree = 4,
stemming=FALSE, labelsize = 10, documents=2)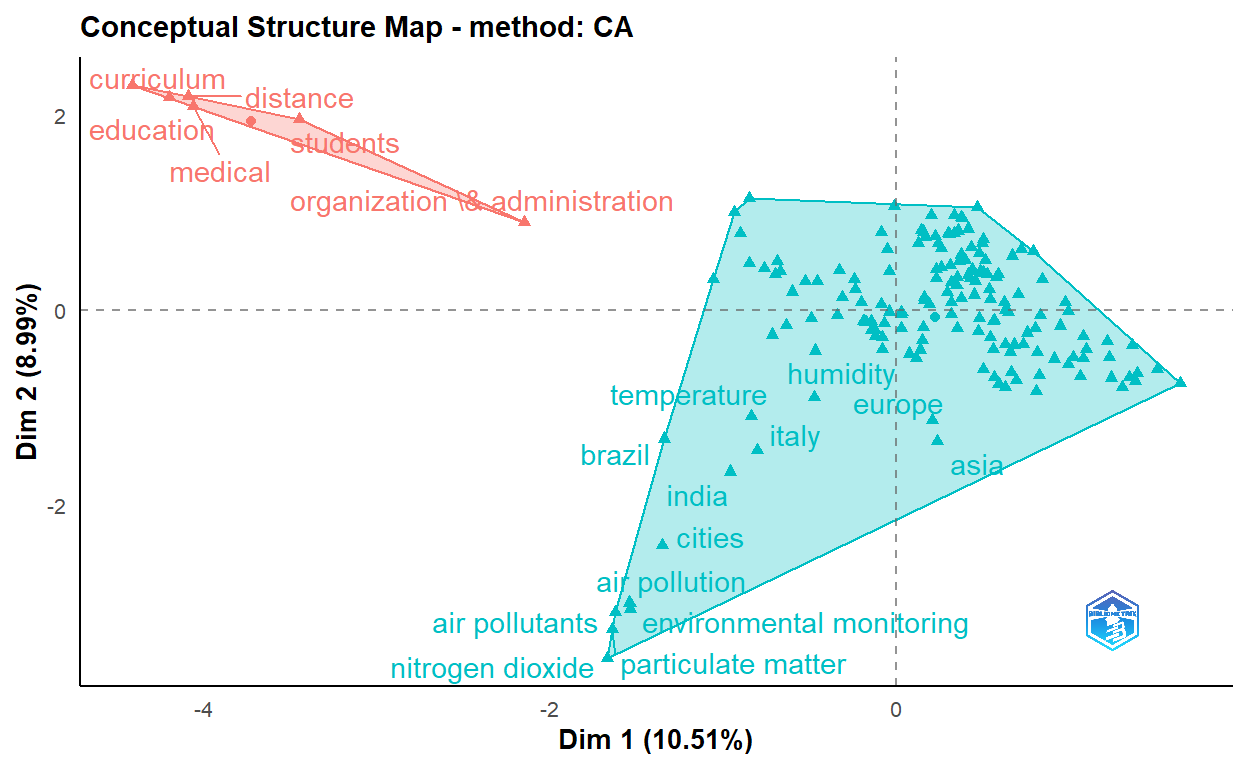
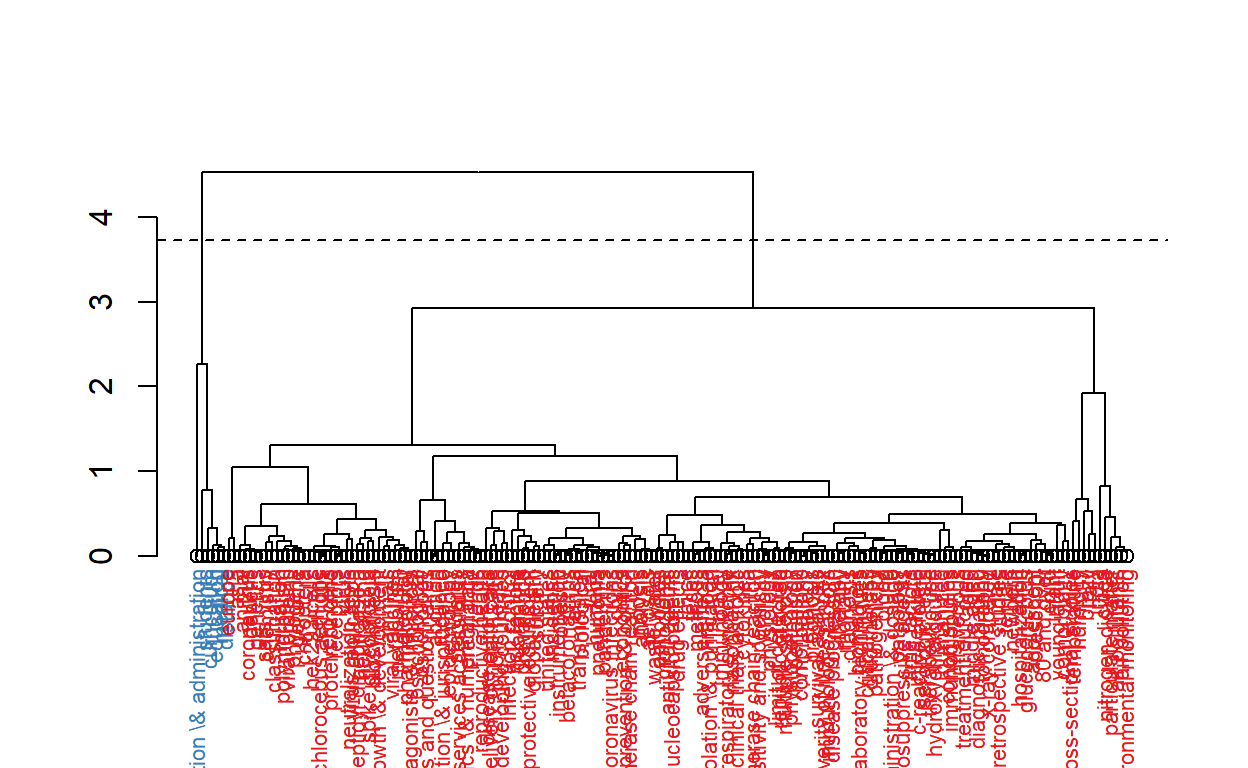
knitr::include_graphics("pic.jpg")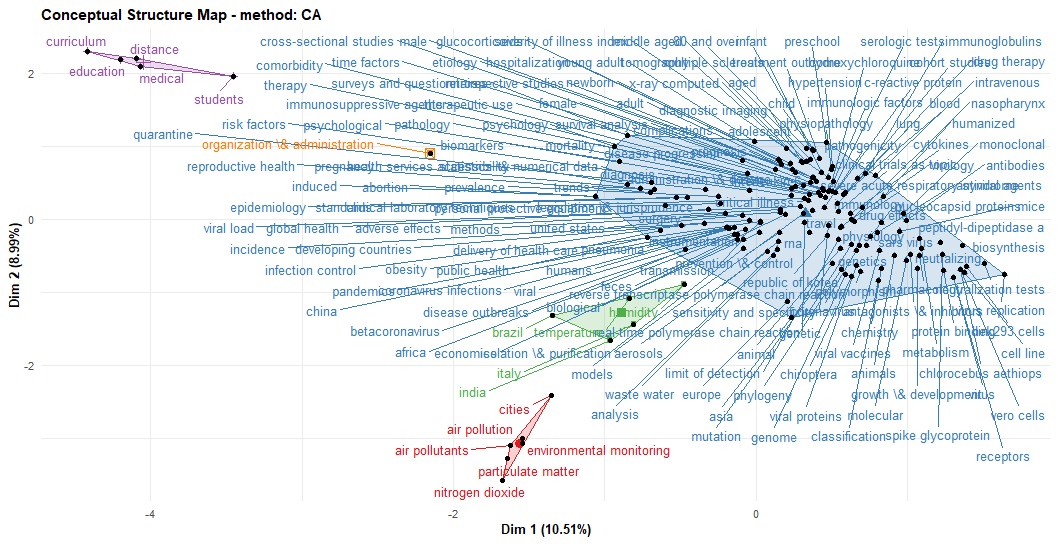
The figure above shows keywords of the articles are clustered into five main categories.
- Clinical, molecular and epidemiological keywords: the biggest part of the cluster with blue text on the figure
- Environmental monitoring cluster
- Humidity studies cluster
- Medical education cluster
- Organization and administration cluster
Now, let’s use bib2df
It is also possible to continue our analysis using the covid19_bibanalysisdata. However, it is good to try another package, bib2df. We will use this package to convert our BibTeX file to data frame and then from there we can perform text mining. It is also good to note that the data frame converted using bibliometrix package may not also work properly with dplyr. But, that depends with your rlang. I have experienced some errors using bibliometrix package with dplyr.
Inspect the data
glimpse(covid19) Rows: 601
Columns: 32
$ CATEGORY <chr> "MISC", "ARTICLE", "ARTICLE", "ARTICLE", "MISC"~
$ BIBTEXKEY <chr> "Gelzinis2020", "Coccia2020", "Ataguba2020", "S~
$ ADDRESS <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ ANNOTE <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ AUTHOR <list> "Gelzinis, Theresa A", "Coccia, Mario", <"Atag~
$ BOOKTITLE <chr> "Journal of cardiothoracic and vascular anesthe~
$ CHAPTER <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ CROSSREF <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ EDITION <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ EDITOR <list> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA~
$ HOWPUBLISHED <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ INSTITUTION <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ JOURNAL <chr> NA, "The Science of the total environment", "Gl~
$ KEY <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ MONTH <chr> "sep", "aug", "dec", "sep", "dec", "nov", "sep"~
$ NOTE <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ NUMBER <chr> "9", NA, "1", NA, "1", NA, NA, "1", NA, NA, "3"~
$ ORGANIZATION <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ PAGES <chr> "2328--2330", "138474", "1788263", "312--321", ~
$ PUBLISHER <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ SCHOOL <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ SERIES <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ TITLE <chr> "Thoracic Anesthesia in the Coronavirus Disease~
$ TYPE <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,~
$ VOLUME <chr> "34", "729", "13", "117", "9", "110", "83", "9"~
$ YEAR <dbl> 2020, 2020, 2020, 2020, 2020, 2020, 2020, 2020,~
$ DOI <chr> "10.1053/j.jvca.2020.05.008", "10.1016/j.scitot~
$ ISSN <chr> "1532-8422 (Electronic)", "1879-1026 (Electroni~
$ LANGUAGE <chr> "eng", "eng", "eng", "eng", "eng", "eng", "eng"~
$ PMID <chr> "32406428", "32498152", "32657669", "32546875",~
$ ABSTRACT <chr> NA, "This study has two goals. The first is to ~
$ KEYWORDS <chr> NA, "Air Pollutants,Air Pollution,Betacoronavir~#Let's choose only some of our variabees
covid19data <- covid19 %>%
select("AUTHOR", "TITLE", "KEYWORDS", "ABSTRACT", "JOURNAL","DOI")
dim(covid19) #601 records, 23 variables[1] 601 32which(!complete.cases(covid19$DOI)) # All of them have missing values[1] 87 551[1] 159[1] 0[1] 2[1] 442table(covid19$CATEGORY) #452 are journal aricles, 149 are Miscelaneous(books, conference abstracst, etc.
ARTICLE MISC
452 149 Let’s filter out the records with no Abstract
#Let's filter the data to retreive only the records with Abstract
#As we have seen above, we have 159 records without abstracts
#use dplyr to filter
library(dplyr)
covid19new <- covid19 %>%
filter(!is.na(ABSTRACT)) #remove all records with missing abstracts
sum(!is.na(covid19new$ABSTRACT)) #442 records have abstracts[1] 442 #BINGO! We excluded the studies without Abstract. Now we, can play with text mining. I am interested only in Journal articles
We need to find out the type of these records: journal articles, books, conference abstracts, etc. We can then focus on journal articles having with abstracts included in PubMed.
table(covid19new$CATEGORY) #382 are journal articles, 60 are Miscelaneous documents
ARTICLE MISC
382 60 # Filter journal articles having abstracts
covid19new <- covid19new %>%
filter(CATEGORY=="ARTICLE")
#Check
table(covid19new$CATEGORY) #382 journal articles
ARTICLE
382 Text mining
- Since we already have our 442 studies with no missing abstract, we can conduct our text mining analysis using this data frame.
- There are several r packages for conducting text mining. Most popular packages are tidytext, text2vec, quanteda, tm and stringr.
- For today, I will simply follow tidytext.
- There are some important steps in text mining such as tokenization, visualization, computing tf-idf statistics, n-grams, etc
- Term frequency inverse document frequency (tf-idf) is a weighted numerical representation of how a certain word is important in a document. It is calculated using the following formula.
\[tfidf( t, d, D ) = tf( t, d ) \times idf( t, D )\] \[idf( t, D ) = log \frac{ \text{| } D \text{ |} }{ 1 + \text{| } \{ d \in D : t \in d \} \text{ |} }\]
Where t is the terms appearing in a document; d denotes each document; D denotes the collection of documents.
Tokenization
- Tokenization is the process of breaking a certain text into word by word columns. For example, if one abstract is written using 300 words, tokenizing the abstract will result in 300 columns for each word. This will make things easy to count words and do any further analysis.
tidy_covid_data <- data %>% unnest_tokens(input=ABSTRACT, output=word)Remove stop words
Stop words are words that are not very relevant to the meaning or concept of the document. For example, see this sentence. “The COVID19 pandemic is the biggest global health crisis of our time”. In this sentence, “the, is, of, our” are not relevant for the concept of this text. These words need to be removed from our analysis. To remove these words, we can use the stopwords data which is available for us. If we want to add additional stop words we can customize and create our own customized stop words.
Customize stop_words
- Usually, abstract contains words like “background”, “introduction”, “materials”, “methods”, “results”, “conclusions”, etc. Let’s remove these words
# since it is all about covid-19, we don't need covid custom_stop_words <- bind_rows(tibble(word=c("covid", "cov", "background", "introduction", "materials", "methods", "results", "conclusions", "0", "1","2", "3", "4", "5", "6", "7", "8", "9","19", "2019", "2020", "95"), lexicon = c("custom")), stop_words) tidy_covid_data <- tidy_covid_data %>% anti_join(custom_stop_words) # count number of words and plot it tidy_covid_data %>% count(word, sort=T) %>% filter(n>100) %>% mutate(word=reorder(word, n)) %>% ggplot(aes(x=word, y=n)) + geom_col(fill="#619CFF") + coord_flip()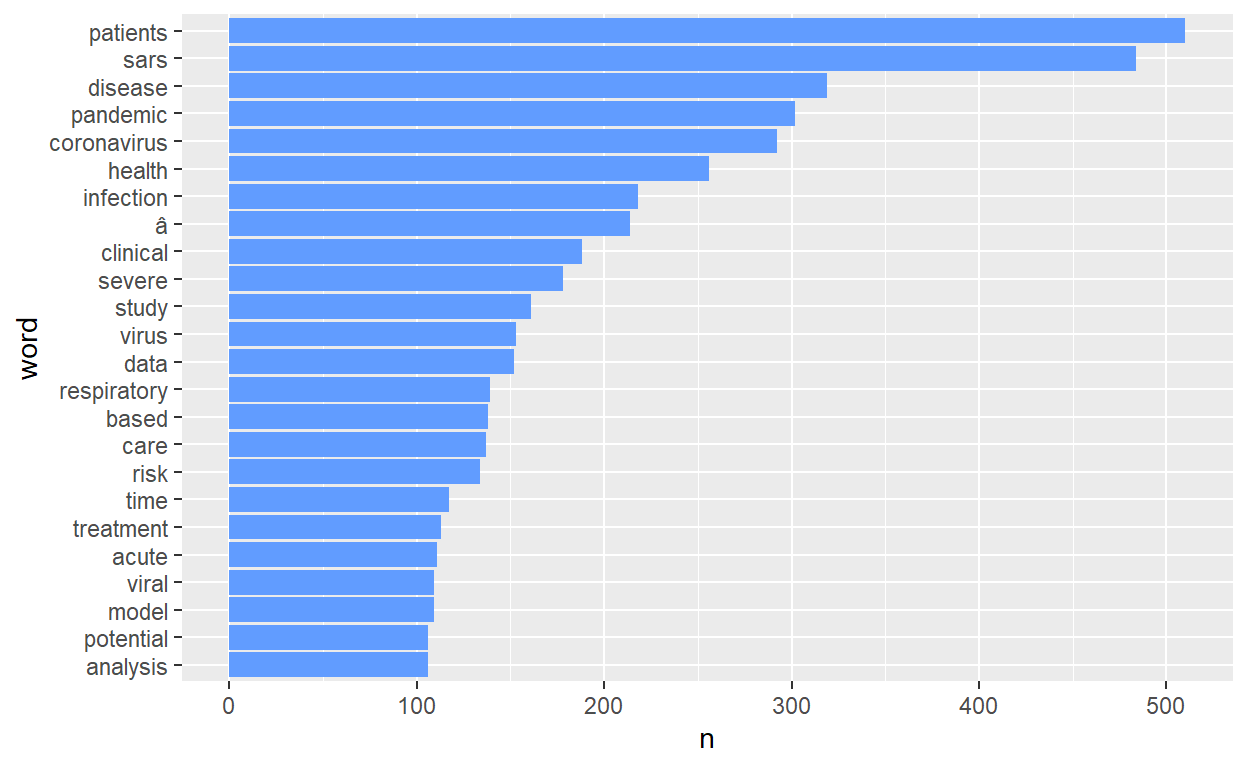
Word cloud
library(wordcloud) pal <- brewer.pal(8, "Dark2") tidy_covid_data %>% count(word) %>% with(wordcloud(word, n, max.words=300, colors = pal))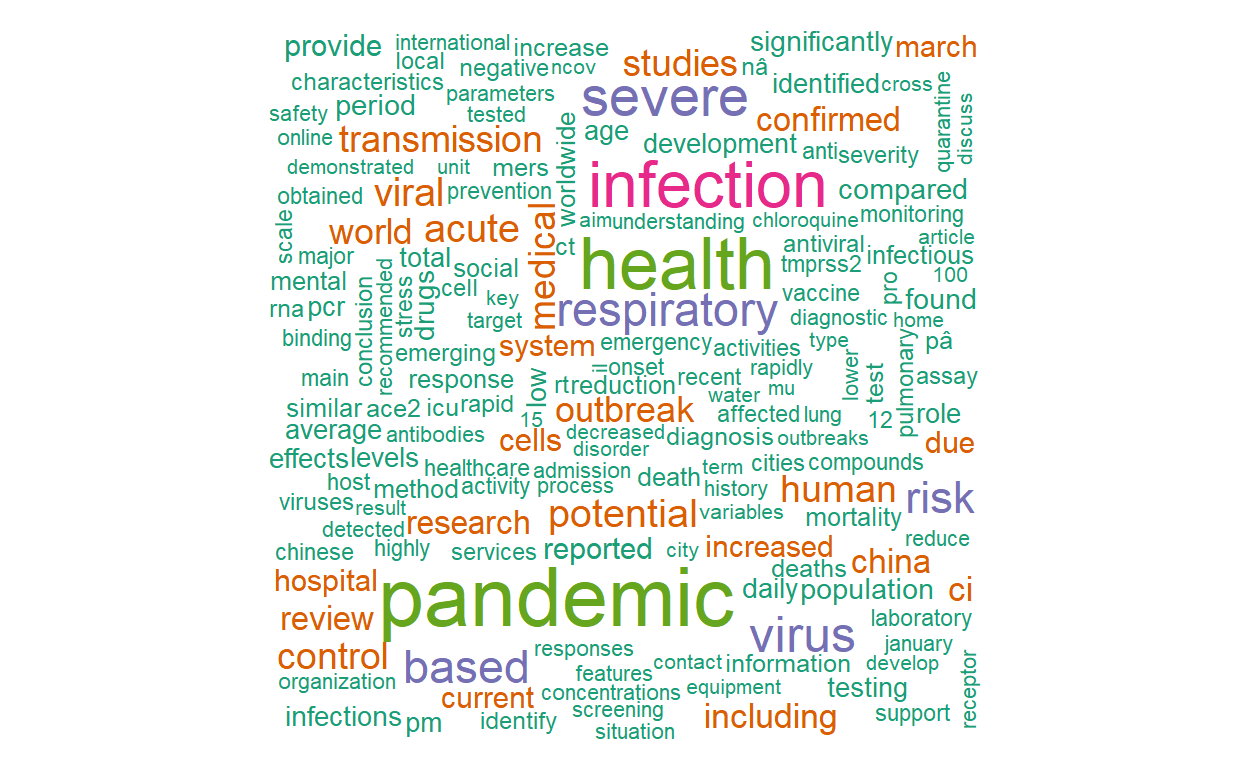
How are words connected to each other in the records?
This needs tokenizing using n-grams. This will be our next stop.
The way forward
R is a powerfull open source tool for systematic reviewers. It is helpful to exclude duplicate records that your reference management software often misses. Currently, I am employing text mining applications to facilitate title/abstract screening, full text screening, clustering of studies and topic modelling. I found text mining is very interesting field and it is really appealing to learn. Looking at the current pace of medical literature, the future of systematic reviews lies on using automated tools, and leveraging text mining and machine learning algorithms. In my opinion, anyone who is interested in systematic reviews needs to consider text mining. The biggest challenge I have so far is, applying machine learning to cluster or classify abstracts is computationally intensive.The curse of high dimensionality!
Contact
Please mention MihiretuKebede1 if you tweet this post.
If you have enjoyed reading this blog post, consider subscribing for upcoming posts.